Protein biosynthesis
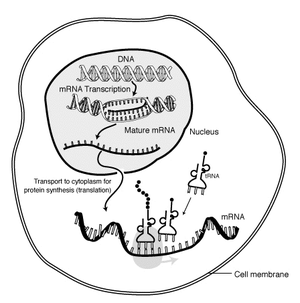
RNA is transcribed in the nucleus; once completely processed, it is transported to the cytoplasm and translated by the ribosome (shown in very pale grey behind the tRNA).
Protein synthesis is the process whereby biological cells generate new proteins; it is balanced by the loss of cellular proteins via degradation or export. Translation, the assembly of amino acids by ribosomes, is an essential part of the biosynthetic pathway, along with generation of messenger RNA (mRNA), aminoacylation of transfer RNA (tRNA), co-translational transport, and post-translational modification. Protein biosynthesis is strictly regulated at multiple steps.[1] They are principally during transcription (phenomena of RNA synthesis from DNA template) and translation (phenomena of amino acid assembly from RNA).
The cistron DNA is transcribed into the first of a series of RNA intermediates. The last version is used as a template in synthesis of a polypeptide chain. Protein will often be synthesized directly from genes by translating mRNA. However, when a protein must be available on short notice or in large quantities, a protein precursor is produced. A proprotein is an inactive protein containing one or more inhibitory peptides that can be activated when the inhibitory sequence is removed by proteolysis during posttranslational modification. A preprotein is a form that contains a signal sequence (an N-terminal signal peptide) that specifies its insertion into or through membranes, i.e., targets them for secretion.[2] The signal peptide is cleaved off in the endoplasmic reticulum.[2]Preproproteins have both sequences (inhibitory and signal) still present.
In protein synthesis, a succession of tRNA molecules charged with appropriate amino acids are brought together with an mRNA molecule and matched up by base-pairing through the anti-codons of the tRNA with successive codons of the mRNA. The amino acids are then linked together to extend the growing protein chain, and the tRNAs, no longer carrying amino acids, are released. This whole complex of processes is carried out by the ribosome, formed of two main chains of RNA, called ribosomal RNA (rRNA), and more than 50 different proteins. The ribosome latches onto the end of an mRNA molecule and moves along it, capturing loaded tRNA molecules and joining together their amino acids to form a new protein chain.[3]
Protein biosynthesis, although very similar, is different for prokaryotes and eukaryotes.
Contents
1 Transcription
2 Translation
3 Events during or following protein translation
4 See also
5 References
6 External links
Transcription

Diagram showing the process of transcription
In transcription an mRNA chain is generated, with one strand of the DNA double helix in the genome as a template. This strand is called the template strand. Transcription can be divided into 3 stages: initiation, elongation, and termination, each regulated by a large number of proteins such as transcription factors and coactivators that ensure that the correct gene is transcribed.
Transcription occurs in the cell nucleus, where the DNA is held and is never able to leave. The DNA structure of the cell is made up of two helixes made up of sugar and phosphate held together by hydrogen bonds between the bases of opposite strands. The sugar and the phosphate in each strand are joined together by stronger phosphodiester covalent bonds. The DNA is "unzipped" (disruption of hydrogen bonds between different single strands) by the enzyme helicase, leaving the single nucleotide chain open to be copied. RNA polymerase reads the DNA strand from the 3-prime (3') end to the 5-prime (5') end, while it synthesizes a single strand of messenger RNA in the 5'-to-3' direction. The general RNA structure is very similar to the DNA structure, but in RNA the nucleotide uracil takes the place that thymine occupies in DNA. The single strand of mRNA leaves the nucleus through nuclear pores, and migrates into the cytoplasm.
The first product of transcription differs in prokaryotic cells from that of eukaryotic cells, as in prokaryotic cells the product is mRNA, which needs no post-transcriptional modification, whereas, in eukaryotic cells, the first product is called primary transcript, that needs post-transcriptional modification (capping with 7-methyl-guanosine, tailing with a poly A tail) to give hnRNA (heterogeneous nuclear RNA). hnRNA then undergoes splicing of introns (noncoding parts of the gene) via spliceosomes to produce the final mRNA.
Translation

Diagram showing the process of translation
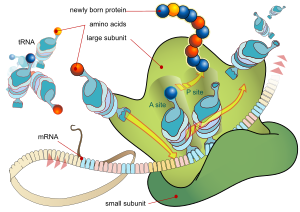
Diagram showing the translation of mRNA and the synthesis of proteins by a ribosome
Phenomena of amino acid assembly from RNA.
The synthesis of proteins from RNA is known as translation. In eukaryotes, translation occurs in the cytoplasm, where the ribosomes are located. Ribosomes are made of a small and large subunit that surround the mRNA. In translation, messenger RNA (mRNA) is decoded to produce a specific polypeptide according to the rules specified by the trinucleotide genetic code. This uses an mRNA sequence as a template to guide the synthesis of a chain of amino acids that form a protein. Translation proceeds in four phases: activation, initiation, elongation, and termination (all describing the growth of the amino acid chain, or polypeptide that is the product of translation).
In activation, the correct amino acid (AA) is joined to the correct transfer RNA (tRNA). While this is not, in the technical sense, a step in translation, it is required for translation to proceed. The AA is joined by its carboxyl group to the 3' OH of the tRNA by an ester bond. When the tRNA has an amino acid linked to it, it is termed "charged".
Initiation involves the small subunit of the ribosome binding to 5' end of mRNA with the help of initiation factors (IF), other proteins that assist the process.
Elongation occurs when the next aminoacyl-tRNA (charged tRNA) in line binds to the ribosome along with GTP and an elongation factor.
Termination of the polypeptide happens when the A site of the ribosome faces a stop codon (UAA, UAG, or UGA). When this happens, no tRNA can recognize it, but releasing factor can recognize nonsense codons and causes the release of the polypeptide chain.
The capacity of disabling or inhibiting translation in protein biosynthesis is used by some antibiotics such as anisomycin, cycloheximide, chloramphenicol, tetracycline, streptomycin, erythromycin, puromycin, etc.
Events during or following protein translation
Events that occur during or following biosynthesis include proteolysis, post-translational modification and protein folding.
Proteolysis may remove N-terminal, C-terminal or internal amino-acid residues or peptides from the polypeptide.
The termini and side-chains of the polypeptide may be subjected to post-translational modification. These modifications may be required for correct cellular localisation or the natural function of the protein.
During and after synthesis, polypeptide chains often fold to assume, so called, native secondary and tertiary structures. This is known as protein folding and is typically required for the natural function of the protein.
See also
- Central dogma of molecular biology
- Cistron
- Gene expression
- Genetic code
- Operon
- Peptide synthesis
- Protein production
References
^ Kafri M, Metzl-Raz E, Jona G, Barkai N. 2016. The Cost of Protein Production. Cell Rep 14:22–31. https://dx.doi.org/10.1016/j.celrep.2015.12.015
^ ab Alberts, Bruce (2002). Molecular biology of the cell. New York: Garland Science. p. 760. ISBN 0-8153-3218-1..mw-parser-output cite.citation{font-style:inherit}.mw-parser-output q{quotes:"""""""'""'"}.mw-parser-output code.cs1-code{color:inherit;background:inherit;border:inherit;padding:inherit}.mw-parser-output .cs1-lock-free a{background:url("//upload.wikimedia.org/wikipedia/commons/thumb/6/65/Lock-green.svg/9px-Lock-green.svg.png")no-repeat;background-position:right .1em center}.mw-parser-output .cs1-lock-limited a,.mw-parser-output .cs1-lock-registration a{background:url("//upload.wikimedia.org/wikipedia/commons/thumb/d/d6/Lock-gray-alt-2.svg/9px-Lock-gray-alt-2.svg.png")no-repeat;background-position:right .1em center}.mw-parser-output .cs1-lock-subscription a{background:url("//upload.wikimedia.org/wikipedia/commons/thumb/a/aa/Lock-red-alt-2.svg/9px-Lock-red-alt-2.svg.png")no-repeat;background-position:right .1em center}.mw-parser-output .cs1-subscription,.mw-parser-output .cs1-registration{color:#555}.mw-parser-output .cs1-subscription span,.mw-parser-output .cs1-registration span{border-bottom:1px dotted;cursor:help}.mw-parser-output .cs1-hidden-error{display:none;font-size:100%}.mw-parser-output .cs1-visible-error{font-size:100%}.mw-parser-output .cs1-subscription,.mw-parser-output .cs1-registration,.mw-parser-output .cs1-format{font-size:95%}.mw-parser-output .cs1-kern-left,.mw-parser-output .cs1-kern-wl-left{padding-left:0.2em}.mw-parser-output .cs1-kern-right,.mw-parser-output .cs1-kern-wl-right{padding-right:0.2em}
^ Alberts, Bruce. Molecular Biology of the Cell, 5e. New York: Garland Science, 2008.
External links
Interactive Java simulation of transcription initiation. From Center for Models of Life at the Niels Bohr Institute.- A website focused primarily on Protein Synthesis.