Potassium channel

Top view of potassium ions (purple) moving through potassium channel (PDB: 1BL8)
Potassium channels are the most widely distributed type of ion channel and are found in virtually all living organisms.[1] They form potassium-selective pores that span cell membranes. Furthermore, potassium channels are found in most cell types and control a wide variety of cell functions.[2][3]
Contents
1 Function
2 Types
3 Structure
3.1 Selectivity filter
3.1.1 Selectivity mechanism
3.2 Hydrophobic region
3.3 Central cavity
4 Regulation
5 Blockers
6 Muscarinic potassium channel
7 In fine art
8 See also
9 References
10 External links
Function
Potassium channels function to conduct potassium ions down their electrochemical gradient, doing so both rapidly (up to the diffusion rate of K+ ions in bulk water) and selectively (excluding, most notably, sodium despite the sub-angstrom difference in ionic radius).[4] Biologically, these channels act to set or reset the resting potential in many cells. In excitable cells, such as neurons, the delayed counterflow of potassium ions shapes the action potential.
By contributing to the regulation of the action potential duration in cardiac muscle, malfunction of potassium channels may cause life-threatening arrhythmias. Potassium channels may also be involved in maintaining vascular tone.
They also regulate cellular processes such as the secretion of hormones (e.g., insulin release from beta-cells in the pancreas) so their malfunction can lead to diseases (such as diabetes).
Types
There are four major classes of potassium channels:
Calcium-activated potassium channel - open in response to the presence of calcium ions or other signalling molecules.
Inwardly rectifying potassium channel - passes current (positive charge) more easily in the inward direction (into the cell).
Tandem pore domain potassium channel - are constitutively open or possess high basal activation, such as the "resting potassium channels" or "leak channels" that set the negative membrane potential of neurons.
Voltage-gated potassium channel - are voltage-gated ion channels that open or close in response to changes in the transmembrane voltage.
The following table contains a comparison of the major classes of potassium channels with representative examples (for a complete list of channels within each class, see the respective class pages).
Class | Subclasses | Function | Blockers | Activators |
|---|---|---|---|---|
Calcium-activated 6T & 1P |
|
|
|
|
Inwardly rectifying 2T & 1P |
|
|
|
|
|
|
|
| |
|
|
|
| |
Tandem pore domain 4T & 2P |
|
|
|
|
Voltage-gated 6T & 1P |
|
|
|
|
Structure

Potassium channel Kv1.2, structure in a membrane-like environment. Calculated hydrocarbon boundaries of the lipid bilayer are indicated by red and blue lines.
Potassium channels have a tetrameric structure in which four identical protein subunits associate to form a fourfold symmetric (C4) complex arranged around a central ion conducting pore (i.e., a homotetramer). Alternatively four related but not identical protein subunits may associate to form heterotetrameric complexes with pseudo C4 symmetry. All potassium channel subunits have a distinctive pore-loop structure that lines the top of the pore and is responsible for potassium selective permeability.
There are over 80 mammalian genes that encode potassium channel subunits. However potassium channels found in bacteria are amongst the most studied of ion channels, in terms of their molecular structure. Using X-ray crystallography,[24][25] profound insights have been gained into how potassium ions pass through these channels and why (smaller) sodium ions do not.[26] The 2003 Nobel Prize for Chemistry was awarded to Rod MacKinnon for his pioneering work in this area.[27]
Selectivity filter
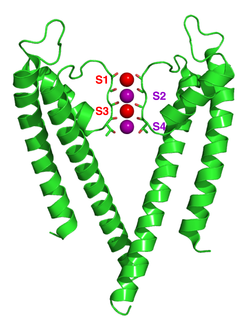
Crystallographic structure of the bacterial KcsA potassium channel (PDB: 1K4C).[28] In this figure, only two of the four subunits of the tetramer are displayed for the sake of clarity. The protein is displayed as a green cartoon diagram. In addition backbone carbonyl groups and threonine sidechain protein atoms (oxygen = red, carbon = green) are displayed. Finally potassium ions (occupying the S2 and S4 sites) and the oxygen atoms of water molecules (S1 and S3) are depicted as purple and red spheres respectively.
Potassium ion channels remove the hydration shell from the ion when it enters the selectivity filter. The selectivity filter is formed by a five residue sequence, TVGYG, termed the signature sequence, within each of the four subunits. This signature sequence is within a loop between the pore helix and TM2/6, historically termed the P-loop. This signature sequence is highly conserved, with the exception that a valine residue in prokaryotic potassium channels is often substituted with an isoleucine residue in eukaryotic channels. This sequence adopts a unique main chain structure, structurally analogous to a nest protein structural motif. The four sets of electronegative carbonyl oxygen atoms are aligned toward the center of the filter pore and form a square anti-prism similar to a water-solvating shell around each potassium binding site. The distance between the carbonyl oxygens and potassium ions in the binding sites of the selectivity filter is the same as between water oxygens in the first hydration shell and a potassium ion in water solution, providing an energetically-favorable route for de-solvation of the ions. This width appears to be maintained by hydrogen bonding and van der Waals forces within a sheet of aromatic amino acid residues surrounding the selectivity filter.[24][29] The selectivity filter opens towards the extracellular solution, exposing four carbonyl oxygens in a glycine residue (Gly79 in KcsA). The next residue toward the extracellular side of the protein is the negatively charged Asp80 (KcsA). This residue together with the five filter residues form the pore that connects the water-filled cavity in the center of the protein with the extracellular solution.[30]
Selectivity mechanism
The mechanism of potassium channel selectivity remains under continued debate. The carbonyl oxygens are strongly electro-negative and cation-attractive. The filter can accommodate potassium ions at 4 sites usually labelled S1 to S4 starting at the extracellular side. In addition, one ion can bind in the cavity at a site called SC or one or more ions at the extracellular side at more or less well-defined sites called S0 or Sext. Several different occupancies of these sites are possible. Since the X-ray structures are averages over many molecules, it is, however, not possible to deduce the actual occupancies directly from such a structure. In general, there is some disadvantage due to electrostatic repulsion to have two neighboring sites occupied by ions. Proposals for the mechanism of selectivity have been made based on molecular dynamics simulations,[31] toy models of ion binding,[32] thermodynamic calculations,[33] topological considerations,[34][35] and structural differences[36] between selective and non-selective channels.
The mechanism for ion translocation in KcsA has been studied extensively by theoretical calculations and simulation.[30][37] The prediction of an ion conduction mechanism in which the two doubly occupied states (S1, S3) and (S2, S4) play an essential role has been affirmed by both techniques. MD simulations suggest the two extracellular states, Sext and S0, reflecting ions entering and leaving the filter, also are important actors in ion conduction.
Hydrophobic region
This region is used to neutralize the environment around the potassium ion so that it is not attracted to any charges. In turn, it speeds up the reaction.
Central cavity
A central pore, 10 Å wide, is located near the center of the transmembrane channel, where the energy barrier is highest for the transversing ion due to the hydrophobity of the channel wall. The water-filled cavity and the polar C-terminus of the pore helices ease the energetic barrier for the ion. Repulsion by preceding multiple potassium ions is thought to aid the throughput of the ions.
The presence of the cavity can be understood intuitively as one of the channel's mechanisms for overcoming the dielectric barrier, or repulsion by the low-dielectric membrane, by keeping the K+ ion in a watery, high-dielectric environment.
Regulation

Graphical representation of open and shut potassium channels (PDB: 1lnq and PDB: 1k4c). Two simple bacterial channels are shown to compare the "open" channel structure on the right with the "closed" structure on the left. At top is the filter (selects potassium ions), and at bottom is the gating domain (controls opening and closing of channel).
The flux of ions through the potassium channel pore is regulated by two related processes, termed gating and inactivation. Gating is the opening or closing of the channel in response to stimuli, while inactivation is the rapid cessation of current from an open potassium channel and the suppression of the channel's ability to resume conducting. While both processes serve to regulate channel conductance, each process may be mediated by a number of mechanisms.
Generally, gating is thought to be mediated by additional structural domains which sense stimuli and in turn open the channel pore. These domains include the RCK domains of BK channels,[38][39][40] and voltage sensor domains of voltage gated K+ channels. These domains are thought to respond to the stimuli by physically opening the intracellular gate of the pore domain, thereby allowing potassium ions to traverse the membrane. Some channels have multiple regulatory domains or accessory proteins, which can act to modulate the response to stimulus. While the mechanisms continue to be debated, there are known structures of a number of these regulatory domains, including RCK domains of prokaryotic[41][42][43] and eukaryotic[38][39][40] channels, pH gating domain of KcsA,[44] cyclic nucleotide gating domains,[45] and voltage gated potassium channels.[46][47]
N-type inactivation is typically the faster inactivation mechanism, and is termed the "ball and chain" model.[48] N-type inactivation involves interaction of the N-terminus of the channel, or an associated protein, which interacts with the pore domain and occludes the ion conduction pathway like a "ball". Alternatively, C-type inactivation is thought to occur within the selectivity filter itself, where structural changes within the filter render it non-conductive. There are a number of structural models of C-type inactivated K+ channel filters,[49][50][51] although the precise mechanism remains unclear.
Blockers
Potassium channel blockers inhibit the flow of potassium ions through the channel. They either compete with potassium binding within the selectivity filter or bind outside the filter to occlude ion conduction. An example of one of these competitors is quaternary ammonium ions, which bind at the extracellular face [52][53] or central cavity of the channel.[54] For blocking from the central cavity quaternary ammonium ions are also known as open channel blockers, as binding classically requires the prior opening of the cytoplasmic gate.[55]
Barium ions can also block potassium channel currents,[56][57] by binding with high affinity within the selectivity filter.[58][59][60][61] This tight binding is thought to underlie barium toxicity by inhibiting potassium channel activity in excitable cells.
Medically potassium channel blockers, such as 4-aminopyridine and 3,4-diaminopyridine, have been investigated for the treatment of conditions such as multiple sclerosis.[62]Off target drug effects can lead to drug induced Long QT syndrome, a potentially life-threatening condition. This is most frequently due to action on the hERG potassium channel in the heart. Accordingly, all new drugs are preclinically tested for cardiac safety.
Muscarinic potassium channel

Birth of an Idea (2007) by Julian Voss-Andreae. The sculpture was commissioned by Roderick MacKinnon based on the molecule's atomic coordinates that were determined by MacKinnon's group in 2001.
Some types of potassium channels are activated by muscarinic receptors and these are called muscarinic potassium channels (IKACh). These channels are a heterotetramer composed of two GIRK1 and two GIRK4 subunits.[63][64] Examples are potassium channels in the heart, which, when activated by parasympathetic signals through M2 muscarinic receptors, cause an outward current of potassium, which slows down the heart rate.[65][66]
In fine art
Roderick MacKinnon commissioned Birth of an Idea, a 5-foot (1.5 m) tall sculpture based on the KcsA potassium channel.[67] The artwork contains a wire object representing the channel's interior with a blown glass object representing the main cavity of the channel structure.
See also
- Inward-rectifier potassium ion channel
- Potassium transporter (Trk) family
- Potassium uptake permease
- Sodium ion channel
- Calcium channel
References
^ Littleton JT, Ganetzky B (Apr 2000). "Ion channels and synaptic organization: analysis of the Drosophila genome". Neuron. 26 (1): 35–43. doi:10.1016/S0896-6273(00)81135-6. PMID 10798390..mw-parser-output cite.citation{font-style:inherit}.mw-parser-output q{quotes:"""""""'""'"}.mw-parser-output code.cs1-code{color:inherit;background:inherit;border:inherit;padding:inherit}.mw-parser-output .cs1-lock-free a{background:url("//upload.wikimedia.org/wikipedia/commons/thumb/6/65/Lock-green.svg/9px-Lock-green.svg.png")no-repeat;background-position:right .1em center}.mw-parser-output .cs1-lock-limited a,.mw-parser-output .cs1-lock-registration a{background:url("//upload.wikimedia.org/wikipedia/commons/thumb/d/d6/Lock-gray-alt-2.svg/9px-Lock-gray-alt-2.svg.png")no-repeat;background-position:right .1em center}.mw-parser-output .cs1-lock-subscription a{background:url("//upload.wikimedia.org/wikipedia/commons/thumb/a/aa/Lock-red-alt-2.svg/9px-Lock-red-alt-2.svg.png")no-repeat;background-position:right .1em center}.mw-parser-output .cs1-subscription,.mw-parser-output .cs1-registration{color:#555}.mw-parser-output .cs1-subscription span,.mw-parser-output .cs1-registration span{border-bottom:1px dotted;cursor:help}.mw-parser-output .cs1-hidden-error{display:none;font-size:100%}.mw-parser-output .cs1-visible-error{font-size:100%}.mw-parser-output .cs1-subscription,.mw-parser-output .cs1-registration,.mw-parser-output .cs1-format{font-size:95%}.mw-parser-output .cs1-kern-left,.mw-parser-output .cs1-kern-wl-left{padding-left:0.2em}.mw-parser-output .cs1-kern-right,.mw-parser-output .cs1-kern-wl-right{padding-right:0.2em}
^ Hille, Bertil (2001). "Chapter 5: Potassium Channels and Chloride Channels". Ion channels of excitable membranes. Sunderland, Mass: Sinauer. pp. 131–168. ISBN 978-0-87893-321-1.
^ Jessell TM, Kandel ER, Schwartz JH (2000). "Chapter 6: Ion Channels". Principles of Neural Science (4th ed.). New York: McGraw-Hill. pp. 105–124. ISBN 978-0-8385-7701-1.
^ Lim, Carmay; Dudev, Todor (2016). "Chapter 10. Potassium Versus Sodium Selectivity in Monovalent Ion Channel Selectivity Filters". In Astrid, Sigel; Helmut, Sigel; Roland K.O., Sigel. The Alkali Metal Ions: Their Role in Life. Metal Ions in Life Sciences. 16. Springer. pp. 325–347. doi:10.1007/978-4-319-21756-7_9 (inactive 2018-12-08).
^ Rang, HP (2003). Pharmacology. Edinburgh: Churchill Livingstone. p. 60. ISBN 978-0-443-07145-4.
^ Kobayashi T, Washiyama K, Ikeda K (Mar 2006). "Inhibition of G protein-activated inwardly rectifying K+ channels by ifenprodil". Neuropsychopharmacology. 31 (3): 516–24. doi:10.1038/sj.npp.1300844. PMID 16123769.
^ abcdef Enyedi P, Czirják G (Apr 2010). "Molecular background of leak K+ currents: two-pore domain potassium channels". Physiological Reviews. 90 (2): 559–605. doi:10.1152/physrev.00029.2009. PMID 20393194.
^ abcdef Lotshaw DP (2007). "Biophysical, pharmacological, and functional characteristics of cloned and native mammalian two-pore domain K+ channels". Cell Biochemistry and Biophysics. 47 (2): 209–56. doi:10.1007/s12013-007-0007-8. PMID 17652773.
^ Fink M, Lesage F, Duprat F, Heurteaux C, Reyes R, Fosset M, Lazdunski M (Jun 1998). "A neuronal two P domain K+ channel stimulated by arachidonic acid and polyunsaturated fatty acids". The EMBO Journal. 17 (12): 3297–308. doi:10.1093/emboj/17.12.3297. PMC 1170668. PMID 9628867.
^ Goldstein SA, Bockenhauer D, O'Kelly I, Zilberberg N (Mar 2001). "Potassium leak channels and the KCNK family of two-P-domain subunits". Nature Reviews. Neuroscience. 2 (3): 175–84. doi:10.1038/35058574. PMID 11256078.
^ Sano Y, Inamura K, Miyake A, Mochizuki S, Kitada C, Yokoi H, Nozawa K, Okada H, Matsushime H, Furuichi K (Jul 2003). "A novel two-pore domain K+ channel, TRESK, is localized in the spinal cord". The Journal of Biological Chemistry. 278 (30): 27406–12. doi:10.1074/jbc.M206810200. PMID 12754259.
^ Czirják G, Tóth ZE, Enyedi P (Apr 2004). "The two-pore domain K+ channel, TRESK, is activated by the cytoplasmic calcium signal through calcineurin". The Journal of Biological Chemistry. 279 (18): 18550–8. doi:10.1074/jbc.M312229200. PMID 14981085.
^ Kindler CH, Yost CS, Gray AT (Apr 1999). "Local anesthetic inhibition of baseline potassium channels with two pore domains in tandem". Anesthesiology. 90 (4): 1092–102. doi:10.1097/00000542-199904000-00024. PMID 10201682.
^ abc Meadows HJ, Randall AD (Mar 2001). "Functional characterisation of human TASK-3, an acid-sensitive two-pore domain potassium channel". Neuropharmacology. 40 (4): 551–9. doi:10.1016/S0028-3908(00)00189-1. PMID 11249964.
^ Kindler CH, Paul M, Zou H, Liu C, Winegar BD, Gray AT, Yost CS (Jul 2003). "Amide local anesthetics potently inhibit the human tandem pore domain background K+ channel TASK-2 (KCNK5)". The Journal of Pharmacology and Experimental Therapeutics. 306 (1): 84–92. doi:10.1124/jpet.103.049809. PMID 12660311.
^ Punke MA, Licher T, Pongs O, Friederich P (Jun 2003). "Inhibition of human TREK-1 channels by bupivacaine". Anesthesia and Analgesia. 96 (6): 1665–73. doi:10.1213/01.ANE.0000062524.90936.1F. PMID 12760993.
^ Lesage F, Guillemare E, Fink M, Duprat F, Lazdunski M, Romey G, Barhanin J (Mar 1996). "TWIK-1, a ubiquitous human weakly inward rectifying K+ channel with a novel structure". The EMBO Journal. 15 (5): 1004–11. PMC 449995. PMID 8605869.
^ Duprat F, Lesage F, Fink M, Reyes R, Heurteaux C, Lazdunski M (Sep 1997). "TASK, a human background K+ channel to sense external pH variations near physiological pH". The EMBO Journal. 16 (17): 5464–71. doi:10.1093/emboj/16.17.5464. PMC 1170177. PMID 9312005.
^ Reyes R, Duprat F, Lesage F, Fink M, Salinas M, Farman N, Lazdunski M (Nov 1998). "Cloning and expression of a novel pH-sensitive two pore domain K+ channel from human kidney". The Journal of Biological Chemistry. 273 (47): 30863–9. doi:10.1074/jbc.273.47.30863. PMID 9812978.
^ Meadows HJ, Benham CD, Cairns W, Gloger I, Jennings C, Medhurst AD, Murdock P, Chapman CG (Apr 2000). "Cloning, localisation and functional expression of the human orthologue of the TREK-1 potassium channel". Pflügers Archiv. 439 (6): 714–22. doi:10.1007/s004240050997. PMID 10784345.
^ Patel AJ, Honoré E, Lesage F, Fink M, Romey G, Lazdunski M (May 1999). "Inhalational anesthetics activate two-pore-domain background K+ channels". Nature Neuroscience. 2 (5): 422–6. doi:10.1038/8084. PMID 10321245.
^ Gray AT, Zhao BB, Kindler CH, Winegar BD, Mazurek MJ, Xu J, Chavez RA, Forsayeth JR, Yost CS (Jun 2000). "Volatile anesthetics activate the human tandem pore domain baseline K+ channel KCNK5". Anesthesiology. 92 (6): 1722–30. doi:10.1097/00000542-200006000-00032. PMID 10839924.
^ Rogawski MA, Bazil CW (Jul 2008). "New molecular targets for antiepileptic drugs: alpha(2)delta, SV2A, and K(v)7/KCNQ/M potassium channels". Current Neurology and Neuroscience Reports. 8 (4): 345–52. doi:10.1007/s11910-008-0053-7. PMC 2587091. PMID 18590620.
^ ab Doyle DA, Morais Cabral J, Pfuetzner RA, Kuo A, Gulbis JM, Cohen SL, Chait BT, MacKinnon R (Apr 1998). "The structure of the potassium channel: molecular basis of K+ conduction and selectivity". Science. 280 (5360): 69–77. Bibcode:1998Sci...280...69D. doi:10.1126/science.280.5360.69. PMID 9525859.
^ MacKinnon R, Cohen SL, Kuo A, Lee A, Chait BT (Apr 1998). "Structural conservation in prokaryotic and eukaryotic potassium channels". Science. 280 (5360): 106–9. Bibcode:1998Sci...280..106M. doi:10.1126/science.280.5360.106. PMID 9525854.
^ Armstrong C (Apr 1998). "The vision of the pore". Science. 280 (5360): 56–7. doi:10.1126/science.280.5360.56. PMID 9556453.
^ "The Nobel Prize in Chemistry 2003". The Nobel Foundation. Retrieved 2007-11-16.
^ Zhou Y, Morais-Cabral JH, Kaufman A, MacKinnon R (2001). "Chemistry of ion coordination and hydration revealed by a K+ channel-Fab complex at 2.0 Â resolution". Nature. 414 (6859): 43–8. Bibcode:2001Natur.414...43Z. doi:10.1038/35102009. PMID 11689936.
^ Sauer, DB; Zeng, W; Raghunathan, S; Jiang, Y (4 October 2011). "Protein interactions central to stabilizing the K+ channel selectivity filter in a four-sited configuration for selective K+ permeation". Proceedings of the National Academy of Sciences of the United States of America. 108 (40): 16634–9. Bibcode:2011PNAS..10816634S. doi:10.1073/pnas.1111688108. PMC 3189067. PMID 21933962.
^ ab Hellgren M, Sandberg L, Edholm O (Mar 2006). "A comparison between two prokaryotic potassium channels (KirBac1.1 and KcsA) in a molecular dynamics (MD) simulation study". Biophysical Chemistry. 120 (1): 1–9. doi:10.1016/j.bpc.2005.10.002. PMID 16253415.
^ Noskov SY, Roux B (Feb 2007). "Importance of hydration and dynamics on the selectivity of the KcsA and NaK channels". The Journal of General Physiology. 129 (2): 135–43. doi:10.1085/jgp.200609633. PMC 2154357. PMID 17227917.
^ Noskov SY, Bernèche S, Roux B (Oct 2004). "Control of ion selectivity in potassium channels by electrostatic and dynamic properties of carbonyl ligands". Nature. 431 (7010): 830–4. Bibcode:2004Natur.431..830N. doi:10.1038/nature02943. PMID 15483608.
^ Varma S, Rempe SB (Aug 2007). "Tuning ion coordination architectures to enable selective partitioning". Biophysical Journal. 93 (4): 1093–9. arXiv:physics/0608180. Bibcode:2007BpJ....93.1093V. doi:10.1529/biophysj.107.107482. PMC 1929028. PMID 17513348.
^ Thomas M, Jayatilaka D, Corry B (Oct 2007). "The predominant role of coordination number in potassium channel selectivity". Biophysical Journal. 93 (8): 2635–43. Bibcode:2007BpJ....93.2635T. doi:10.1529/biophysj.107.108167. PMC 1989715. PMID 17573427.
^ Bostick DL, Brooks CL (May 2007). "Selectivity in K+ channels is due to topological control of the permeant ion's coordinated state". Proceedings of the National Academy of Sciences of the United States of America. 104 (22): 9260–5. Bibcode:2007PNAS..104.9260B. doi:10.1073/pnas.0700554104. PMC 1890482. PMID 17519335.
^ Derebe MG, Sauer DB, Zeng W, Alam A, Shi N, Jiang Y (Jan 2011). "Tuning the ion selectivity of tetrameric cation channels by changing the number of ion binding sites". Proceedings of the National Academy of Sciences of the United States of America. 108 (2): 598–602. Bibcode:2011PNAS..108..598D. doi:10.1073/pnas.1013636108. PMC 3021048. PMID 21187421.
^ Morais-Cabral JH, Zhou Y, MacKinnon R (Nov 2001). "Energetic optimization of ion conduction rate by the K+ selectivity filter". Nature. 414 (6859): 37–42. Bibcode:2001Natur.414...37M. doi:10.1038/35102000. PMID 11689935.
^ ab Yuan P, Leonetti MD, Pico AR, Hsiung Y, MacKinnon R (Jul 2010). "Structure of the human BK channel Ca2+-activation apparatus at 3.0 A resolution". Science. 329 (5988): 182–6. Bibcode:2010Sci...329..182Y. doi:10.1126/science.1190414. PMC 3022345. PMID 20508092.
^ ab Wu Y, Yang Y, Ye S, Jiang Y (Jul 2010). "Structure of the gating ring from the human large-conductance Ca(2+)-gated K(+) channel". Nature. 466 (7304): 393–7. Bibcode:2010Natur.466..393W. doi:10.1038/nature09252. PMC 2910425. PMID 20574420.
^ ab Jiang Y, Pico A, Cadene M, Chait BT, MacKinnon R (Mar 2001). "Structure of the RCK domain from the E. coli K+ channel and demonstration of its presence in the human BK channel". Neuron. 29 (3): 593–601. doi:10.1016/S0896-6273(01)00236-7. PMID 11301020.
^ Jiang Y, Lee A, Chen J, Cadene M, Chait BT, MacKinnon R (May 2002). "Crystal structure and mechanism of a calcium-gated potassium channel". Nature. 417 (6888): 515–22. Bibcode:2002Natur.417..515J. doi:10.1038/417515a. PMID 12037559.
^ Kong C, Zeng W, Ye S, Chen L, Sauer DB, Lam Y, Derebe MG, Jiang Y (2012). "Distinct gating mechanisms revealed by the structures of a multi-ligand gated K(+) channel". eLife. 1: e00184. doi:10.7554/eLife.00184. PMC 3510474. PMID 23240087.
^ Cao Y, Jin X, Huang H, Derebe MG, Levin EJ, Kabaleeswaran V, Pan Y, Punta M, Love J, Weng J, Quick M, Ye S, Kloss B, Bruni R, Martinez-Hackert E, Hendrickson WA, Rost B, Javitch JA, Rajashankar KR, Jiang Y, Zhou M (Mar 2011). "Crystal structure of a potassium ion transporter, TrkH". Nature. 471 (7338): 336–40. Bibcode:2011Natur.471..336C. doi:10.1038/nature09731. PMC 3077569. PMID 21317882.
^ Uysal S, Cuello LG, Cortes DM, Koide S, Kossiakoff AA, Perozo E (Jul 2011). "Mechanism of activation gating in the full-length KcsA K+ channel". Proceedings of the National Academy of Sciences of the United States of America. 108 (29): 11896–9. Bibcode:2011PNAS..10811896U. doi:10.1073/pnas.1105112108. PMC 3141920. PMID 21730186.
^ Clayton GM, Silverman WR, Heginbotham L, Morais-Cabral JH (Nov 2004). "Structural basis of ligand activation in a cyclic nucleotide regulated potassium channel". Cell. 119 (5): 615–27. doi:10.1016/j.cell.2004.10.030. PMID 15550244.
^ Jiang Y, Lee A, Chen J, Ruta V, Cadene M, Chait BT, MacKinnon R (May 2003). "X-ray structure of a voltage-dependent K+ channel". Nature. 423 (6935): 33–41. Bibcode:2003Natur.423...33J. doi:10.1038/nature01580. PMID 12721618.
^ Long SB, Campbell EB, Mackinnon R (Aug 2005). "Crystal structure of a mammalian voltage-dependent Shaker family K+ channel". Science. 309 (5736): 897–903. Bibcode:2005Sci...309..897L. doi:10.1126/science.1116269. PMID 16002581.
^ Antz C, Fakler B (Aug 1998). "Fast Inactivation of Voltage-Gated K(+) Channels: From Cartoon to Structure". News in Physiological Sciences. 13: 177–182. PMID 11390785.
^ Cheng WW, McCoy JG, Thompson AN, Nichols CG, Nimigean CM (Mar 2011). "Mechanism for selectivity-inactivation coupling in KcsA potassium channels". Proceedings of the National Academy of Sciences of the United States of America. 108 (13): 5272–7. Bibcode:2011PNAS..108.5272C. doi:10.1073/pnas.1014186108. PMC 3069191. PMID 21402935.
^ Cuello LG, Jogini V, Cortes DM, Perozo E (Jul 2010). "Structural mechanism of C-type inactivation in K(+) channels". Nature. 466 (7303): 203–8. Bibcode:2010Natur.466..203C. doi:10.1038/nature09153. PMC 3033749. PMID 20613835.
^ Cuello LG, Jogini V, Cortes DM, Pan AC, Gagnon DG, Dalmas O, Cordero-Morales JF, Chakrapani S, Roux B, Perozo E (Jul 2010). "Structural basis for the coupling between activation and inactivation gates in K(+) channels". Nature. 466 (7303): 272–5. Bibcode:2010Natur.466..272C. doi:10.1038/nature09136. PMC 3033755. PMID 20613845.
^ Luzhkov VB, Aqvist J (Feb 2005). "Ions and blockers in potassium channels: insights from free energy simulations". Biochimica et Biophysica Acta. 1747 (1): 109–120. doi:10.1016/j.bbapap.2004.10.006. PMID 15680245.
^ Luzhkov VB, Osterberg F, Aqvist J (Nov 2003). "Structure-activity relationship for extracellular block of K+ channels by tetraalkylammonium ions". FEBS Letters. 554 (1–2): 159–164. doi:10.1016/S0014-5793(03)01117-7. PMID 14596932.
^ Posson DJ, McCoy JG, Nimigean CM (Feb 2013). "The voltage-dependent gate in MthK potassium channels is located at the selectivity filter". Nature Structural & Molecular Biology. 20 (2): 159–66. doi:10.1038/nsmb.2473. PMC 3565016. PMID 23262489.
^ Choi KL, Mossman C, Aubé J, Yellen G (Mar 1993). "The internal quaternary ammonium receptor site of Shaker potassium channels". Neuron. 10 (3): 533–41. doi:10.1016/0896-6273(93)90340-w. PMID 8461140.
^ Piasta KN, Theobald DL, Miller C (Oct 2011). "Potassium-selective block of barium permeation through single KcsA channels". The Journal of General Physiology. 138 (4): 421–36. doi:10.1085/jgp.201110684. PMC 3182450. PMID 21911483.
^ Neyton J, Miller C (Nov 1988). "Potassium blocks barium permeation through a calcium-activated potassium channel". The Journal of General Physiology. 92 (5): 549–67. doi:10.1085/jgp.92.5.549. PMC 2228918. PMID 3235973.
^ Lockless SW, Zhou M, MacKinnon R (May 2007). "Structural and thermodynamic properties of selective ion binding in a K+ channel". PLoS Biology. 5 (5): e121. doi:10.1371/journal.pbio.0050121. PMC 1858713. PMID 17472437.
^ Jiang Y, MacKinnon R (Mar 2000). "The barium site in a potassium channel by x-ray crystallography". The Journal of General Physiology. 115 (3): 269–72. doi:10.1085/jgp.115.3.269. PMC 2217209. PMID 10694255.
^ Lam YL, Zeng W, Sauer DB, Jiang Y (Aug 2014). "The conserved potassium channel filter can have distinct ion binding profiles: structural analysis of rubidium, cesium, and barium binding in NaK2K". The Journal of General Physiology. 144 (2): 181–92. doi:10.1085/jgp.201411191. PMC 4113894. PMID 25024267.
^ Guo R, Zeng W, Cui H, Chen L, Ye S (Aug 2014). "Ionic interactions of Ba2+ blockades in the MthK K+ channel". The Journal of General Physiology. 144 (2): 193–200. doi:10.1085/jgp.201411192. PMC 4113901. PMID 25024268.
^ Judge SI, Bever CT (Jul 2006). "Potassium channel blockers in multiple sclerosis: neuronal Kv channels and effects of symptomatic treatment". Pharmacology & Therapeutics. 111 (1): 224–59. doi:10.1016/j.pharmthera.2005.10.006. PMID 16472864.
^ Krapivinsky G, Gordon EA, Wickman K, Velimirović B, Krapivinsky L, Clapham DE (Mar 1995). "The G-protein-gated atrial K+ channel IKACh is a heteromultimer of two inwardly rectifying K(+)-channel proteins". Nature. 374 (6518): 135–41. Bibcode:1995Natur.374..135K. doi:10.1038/374135a0. PMID 7877685.
^ Corey S, Krapivinsky G, Krapivinsky L, Clapham DE (Feb 1998). "Number and stoichiometry of subunits in the native atrial G-protein-gated K+ channel, IKACh". The Journal of Biological Chemistry. 273 (9): 5271–8. doi:10.1074/jbc.273.9.5271. PMID 9478984.
^ Kunkel MT, Peralta EG (Nov 1995). "Identification of domains conferring G protein regulation on inward rectifier potassium channels". Cell. 83 (3): 443–9. doi:10.1016/0092-8674(95)90122-1. PMID 8521474.
^ Wickman K, Krapivinsky G, Corey S, Kennedy M, Nemec J, Medina I, Clapham DE (Apr 1999). "Structure, G protein activation, and functional relevance of the cardiac G protein-gated K+ channel, IKACh". Annals of the New York Academy of Sciences. 868 (1): 386–98. Bibcode:1999NYASA.868..386W. doi:10.1111/j.1749-6632.1999.tb11300.x. PMID 10414308. Archived from the original on 2006-01-29.
^ Ball P (March 2008). "The crucible: Art inspired by science should be more than just a pretty picture". Chemistry World. 5 (3): 42–43. Retrieved 2009-01-12.
External links
Proteopedia channel Potassium channel in 3D
Potassium+Channels at the US National Library of Medicine Medical Subject Headings (MeSH)
Neuromuscular Disease Center (2008-03-04). "Potassium Channels". Washington University in St. Louis. Retrieved 2008-03-10.