Lactobacillus
Lactobacillus
Jump to navigation
Jump to search
This article needs additional citations for verification. (March 2019) (Learn how and when to remove this template message) |
Lactobacillus | |
|---|---|
 | |
Lactobacillus sp. near a squamous epithelial cell | |
Scientific classification | |
| Domain: | Bacteria |
| Phylum: | Firmicutes |
| Class: | Bacilli |
| Order: | Lactobacillales |
| Family: | Lactobacillaceae |
| Genus: | Lactobacillus Beijerinck 1901 |
Species and subspecies[1] | |
| |
Lactobacillus is a genus of Gram-positive, facultative anaerobic or microaerophilic, rod-shaped, non-spore-forming bacteria.[2] They are a major part of the lactic acid bacteria group (i.e., they convert sugars to lactic acid). In humans, they constitute a significant component of the microbiota at a number of body sites, such as the digestive system, urinary system, and genital system. In women of European ancestry, Lactobacillus species are normally a major part of the vaginal microbiota.[3][4]Lactobacillus forms biofilms in the vaginal and gut microbiota, allowing them to persist during harsh environmental conditions and maintain ample populations.[5]Lactobacillus exhibits a mutualistic relationship with the human body, as it protects the host against potential invasions by pathogens, and in turn, the host provides a source of nutrients.[6]Lactobacillus is the most common probiotic found in food such as yogurt, and it is diverse in its application to maintain human well-being, as it can help treat diarrhea, vaginal infections, and skin disorders such as eczema.[7]
Contents
1 Metabolism
2 Genomes
3 Taxonomy
4 Human health
4.1 Vaginal tract
4.2 Interactions with other pathogens
4.3 Probiotics
4.4 Oral health
5 Food production
6 See also
7 References
8 External links
Metabolism[edit]
This section needs additional citations for verification. (April 2015) (Learn how and when to remove this template message) |
Many lactobacilli operate using homofermentative metabolism (they produce only lactic acid from sugars), and some species use heterofermentative metabolism (they can produce either alcohol or lactic acid from sugars).[8] They are aerotolerant despite the complete absence of a respiratory chain.[9][10] This aerotolerance is manganese-dependent and has been explored (and explained) in Lactobacillus plantarum.[9] Many species of this genus do not require iron for growth and have an extremely high hydrogen peroxide tolerance.[citation needed]
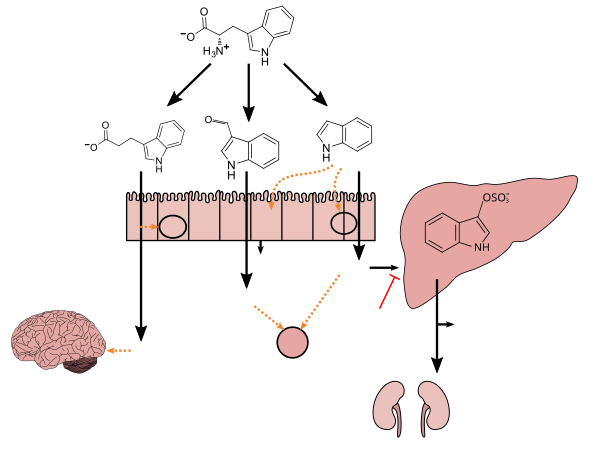
Tryptophan metabolism by human gastrointestinal microbiota ( ) |
Genomes[edit]
The genomes of Lactobacillus are highly variable, ranging in size from 1.2 to 4.9 Mb (megabases). Accordingly, the number of protein-coding genes ranges from 1,267 to about 4,758 genes (in L. sanfranciscensis and L. parakefiri, respectively).[15][16] Even within a single species there can be substantial variation. For instance, strains of L. crispatus have genome sizes ranging from 1.83 to 2.7 Mb, or 1,839 to 2,688 open reading frames.[17]
Lactobacillus contains a wealth of compound microsatellites in the coding region of the genome, which are imperfect and have variant motifs.[18]
Taxonomy[edit]
The genus Lactobacillus currently contains over 180 species and encompasses a wide variety of organisms.[19] The genus is polyphyletic, with the genus Pediococcus dividing the L. casei group, and the species L. acidophilus, L. salivarius, and L. reuteri being representatives of three distinct subclades. The genus Paralactobacillus falls within the L. salivarius group. In recent years, other members of the genus Lactobacillus (formerly known as the Leuconostoc branch of Lactobacillus) have been reclassified into the genera Atopobium, Carnobacterium, Weissella, Oenococcus, and Leuconostoc. More recently, the Pediococcus species P. dextrinicus has been reclassified as a Lactobacillus species.[20] According to metabolism, Lactobacillus species can be divided into three groups:
- Obligately homofermentative (group I) including:
L. acidophilus, L. delbrueckii, L. helveticus, L. salivarius
- Facultatively heterofermentative (group II) including:
L. casei, L. curvatus, L. plantarum, L. sakei
- Obligately heterofermentative (group III) including:
L. brevis, L. buchneri, L. fermentum, L. reuteri
Human health[edit]
Vaginal tract[edit]
The female genital tract is one of the principal colonisation sites for human microbiota, and there is interest in the relationship between the composition of these bacteria and human health, with a domination by a single species being correlated with general welfare and good outcomes in pregnancy. In around 70% of women, a Lactobacillus species is dominant, although that has been found to vary between American women of European origin and those of African origin, the latter group tending to have more diverse vaginal microbiota. Similar differences have also been identified in comparisons between Belgian and Tanzanian women.[3]
Interactions with other pathogens[edit]
Lactobacillus species produce hydrogen peroxide which inhibits the growth and virulence of the fungal pathogen Candida albicans in vitro and in vivo.[21][22] In vitro studies have also shown that Lactobacillus sp. reduce the pathogenicity of C. albicans through the production of organic acids and certain metabolites.[23] Both the presence of metabolites, such as sodium butyrate, and the decrease in environmental pH caused by the organic acids reduce the growth of hypha in C. albicans, which reduces its pathogenicity.[23]Lactobacillus sp. also reduce the pathogenicity of C. albicans by reducing C. albicans biofilm formation.[23] Biofilm formation is reduced by both the competition from Lactobacillus sp., and the formation of defective biofilms which is linked to the reduced hypha growth mentioned earlier.[23] On the other hand, following antibiotic therapy, certain Candida species can suppress the regrowth of Lactobacillus sp. at body sites where they cohabitate, such as in the gastrointestinal tract.[21][22]
In addition to its effects on C. albicans, Lactobacillus sp. also interact with other pathogens. For example, Lactobacillus reuteri can inhibit the growth of many different bacterial species by using glycerol to produce the antimicrobial substance called reuterin.[24] Another example is Lactobacillus salivarius, which interacts with many pathogens through the production of salivaricin B, a bacteriocin.[25]
Probiotics[edit]
Lactobacillus species administered in combination with other probiotics benefits cases of irritable bowel syndrome (IBS), although the extent of efficacy is still uncertain.[26] The probiotics help treat IBS by returning homeostasis when the gut microbiota experiences unusually high levels of opportunistic bacteria.[6] In addition, Lactobacillus species can be administered as probiotics during cases of infection by the ulcer-causing bacterium Helicobacter pylori.[27] Helicobacter pylori is linked to cancer, and antibiotic resistance impedes the success of current antibiotic-based eradication treatments.[27] When Lactobacillus probiotics are administered along with the treatment as an adjuvant, its efficacy is substantially increased and side effects may be lessened.[27] Also, Lactobacillus is used to help control urogenital and vaginal infections, such as bacterial vaginosis (BV). Lactobacillus produce bacteriocins to suppress pathogenic growth of certain bacteria,[28] as well as lactic acid and H2O2 (hydrogen peroxide). Lactic acid lowers the vaginal pH to around 4.5 or less, hampering the survival of other bacteria, and H2O2 reestablishes the normal bacterial microbiota and normal vaginal pH.[28] In children, Lactobacillus strains such as L. rhamnosus are associated with a reduction of atopic eczema, also known as dermatitis, due to anti-inflammatory cytokines secreted by this probiotic bacteria.[6].In addition, lactobacillus with other probiotic[29] organisms in ripen milk and yogurt progress the immunity in the mucous intestine in humans through rising the number of LgA (+)
Oral health[edit]

Dental caries
Some Lactobacillus species have been associated with cases of dental caries (cavities). Lactic acid can corrode teeth, and the Lactobacillus count in saliva has been used as a "caries test" for many years. Lactobacilli characteristically cause existing carious lesions to progress, especially those in coronal caries. The issue is, however, complex, as recent studies show probiotics can allow beneficial lactobacilli to populate sites on teeth, preventing streptococcal pathogens from taking hold and inducing dental decay. The scientific research of lactobacilli in relation to oral health is a new field and only a few studies and results have been published.[30][31] Some studies have provided evidence of certain Lactobacilli which can be a probiotic for oral health.[32] Some species, but not all, show evidence in defense to dental caries.[32] Due to these studies, there have been applications of incorporating such probiotics in chewing gum and lozenges.[32] There is also evidence of certain Lactobacilli that are beneficial in the defense of periodontal disease such as gingivitis and periodontitis.[32]
Food production[edit]
Some Lactobacillus species are used as starter cultures in industry for controlled fermentation in the production of wine, yogurt, cheese, sauerkraut, pickles, beer, cider, kimchi, cocoa, kefir, and other fermented foods, as well as animal feeds and the bokashi soil amendment. The antibacterial and antifungal activity of Lactobacillus species rely on production of bacteriocins and low molecular weight compounds that inhibits these microorganisms.[33][34]
Sourdough bread is made either spontaneously, by taking advantage of the bacteria naturally present in flour, or by using a "starter culture", which is a symbiotic culture of yeast and lactic acid bacteria growing in a water and flour medium. The bacteria metabolize sugars into lactic acid, which lowers the pH of their environment, creating a signature "sourness" associated with yogurt, sauerkraut, etc.
In many traditional pickling processes, vegetables are submerged in brine, and salt-tolerant Lactobacillus species feed on natural sugars found in the vegetables. The resulting mix of salt and lactic acid is a hostile environment for other microbes, such as fungi, and the vegetables are thus preserved—remaining edible for long periods.
Lactobacilli, especially L. casei and L. brevis, are some of the most common beer spoilage organisms. They are, however, essential to the production of sour beers such as Belgian lambics and American wild ales, giving the beer a distinct tart flavor.
See also[edit]
- Lactobacillus L. anticaries
- Lactic acid fermentation
- MRS agar
- Pediococcus
- Probiotics
- Proteobiotics
- Carbon monoxide-releasing molecules
References[edit]
^ "Lactobacillus" (HTML). NCBI taxonomy. Bethesda, MD: National Center for Biotechnology Information. Retrieved 1 March 2019..mw-parser-output cite.citation{font-style:inherit}.mw-parser-output .citation q{quotes:"""""""'""'"}.mw-parser-output .citation .cs1-lock-free a{background:url("//upload.wikimedia.org/wikipedia/commons/thumb/6/65/Lock-green.svg/9px-Lock-green.svg.png")no-repeat;background-position:right .1em center}.mw-parser-output .citation .cs1-lock-limited a,.mw-parser-output .citation .cs1-lock-registration a{background:url("//upload.wikimedia.org/wikipedia/commons/thumb/d/d6/Lock-gray-alt-2.svg/9px-Lock-gray-alt-2.svg.png")no-repeat;background-position:right .1em center}.mw-parser-output .citation .cs1-lock-subscription a{background:url("//upload.wikimedia.org/wikipedia/commons/thumb/a/aa/Lock-red-alt-2.svg/9px-Lock-red-alt-2.svg.png")no-repeat;background-position:right .1em center}.mw-parser-output .cs1-subscription,.mw-parser-output .cs1-registration{color:#555}.mw-parser-output .cs1-subscription span,.mw-parser-output .cs1-registration span{border-bottom:1px dotted;cursor:help}.mw-parser-output .cs1-ws-icon a{background:url("//upload.wikimedia.org/wikipedia/commons/thumb/4/4c/Wikisource-logo.svg/12px-Wikisource-logo.svg.png")no-repeat;background-position:right .1em center}.mw-parser-output code.cs1-code{color:inherit;background:inherit;border:inherit;padding:inherit}.mw-parser-output .cs1-hidden-error{display:none;font-size:100%}.mw-parser-output .cs1-visible-error{font-size:100%}.mw-parser-output .cs1-maint{display:none;color:#33aa33;margin-left:0.3em}.mw-parser-output .cs1-subscription,.mw-parser-output .cs1-registration,.mw-parser-output .cs1-format{font-size:95%}.mw-parser-output .cs1-kern-left,.mw-parser-output .cs1-kern-wl-left{padding-left:0.2em}.mw-parser-output .cs1-kern-right,.mw-parser-output .cs1-kern-wl-right{padding-right:0.2em}
^ Makarova K, Slesarev A, Wolf Y, Sorokin A, Mirkin B, Koonin E, et al. (October 2006). "Comparative genomics of the lactic acid bacteria". Proceedings of the National Academy of Sciences of the United States of America. 103 (42): 15611–6. Bibcode:2006PNAS..10315611M. doi:10.1073/pnas.0607117103. PMC 1622870. PMID 17030793.
^ ab Ma B, Forney LJ, Ravel J (20 September 2012). "Vaginal microbiome: rethinking health and disease". Annual Review of Microbiology. 66 (1): 371–89. doi:10.1146/annurev-micro-092611-150157. PMC 3780402. PMID 22746335.
^ Fettweis JM, Brooks JP, Serrano MG, Sheth NU, Girerd PH, Edwards DJ, Strauss JF, Jefferson KK, Buck GA (October 2014). "Differences in vaginal microbiome in African American women versus women of European ancestry". Microbiology. 160 (Pt 10): 2272–2282. doi:10.1099/mic.0.081034-0. PMC 4178329. PMID 25073854.
^ Salas-Jara MJ, Ilabaca A, Vega M, García A (September 2016). "Biofilm Forming Lactobacillus: New Challenges for the Development of Probiotics". Microorganisms. 4 (3): 35. doi:10.3390/microorganisms4030035. PMC 5039595. PMID 27681929.
^ abc Martín R, Miquel S, Ulmer J, Kechaou N, Langella P, Bermúdez-Humarán LG (July 2013). "Role of commensal and probiotic bacteria in human health: a focus on inflammatory bowel disease". Microbial Cell Factories. 12 (71): 71. doi:10.1186/1475-2859-12-71. PMC 3726476. PMID 23876056.
^ Inglin R. Combined Phenotypic-Genotypic Analyses of the Genus Lactobacillus and Selection of Cultures for Biopreservation of Fermented Food. ETHZ research collection (Ph.D. thesis). ETH Zurich. doi:10.3929/ethz-b-000214904.
^ Zaunmüller T, Eichert M, Richter H, Unden G (September 2006). "Variations in the energy metabolism of biotechnologically relevant heterofermentative lactic acid bacteria during growth on sugars and organic acids". Applied Microbiology and Biotechnology. 72 (3): 421–9. doi:10.1007/s00253-006-0514-3. PMID 16826375.
^ ab Archibald FS, Fridovich I (June 1981). "Manganese, superoxide dismutase, and oxygen tolerance in some lactic acid bacteria". Journal of Bacteriology. 146 (3): 928–36. PMC 216946. PMID 6263860.
^ Small PL, Waterman SR (June 1998). "Acid stress, anaerobiosis and gadCB: lessons from Lactococcus lactis and Escherichia coli". Trends in Microbiology. 6 (6): 214–6. doi:10.1016/S0966-842X(98)01285-2. PMID 9675796.
^ abcdefghi Zhang LS, Davies SS (April 2016). "Microbial metabolism of dietary components to bioactive metabolites: opportunities for new therapeutic interventions". Genome Med. 8 (1): 46. doi:10.1186/s13073-016-0296-x. PMC 4840492. PMID 27102537.Lactobacillus spp. convert tryptophan to indole-3-aldehyde (I3A) through unidentified enzymes [125]. Clostridium sporogenes convert tryptophan to IPA [6], likely via a tryptophan deaminase. ... IPA also potently scavenges hydroxyl radicals
Table 2: Microbial metabolites: their synthesis, mechanisms of action, and effects on health and disease
Figure 1: Molecular mechanisms of action of indole and its metabolites on host physiology and disease
^ Wikoff WR, Anfora AT, Liu J, Schultz PG, Lesley SA, Peters EC, Siuzdak G (March 2009). "Metabolomics analysis reveals large effects of gut microflora on mammalian blood metabolites". Proc. Natl. Acad. Sci. U.S.A. 106 (10): 3698–3703. doi:10.1073/pnas.0812874106. PMC 2656143. PMID 19234110.Production of IPA was shown to be completely dependent on the presence of gut microflora and could be established by colonization with the bacterium Clostridium sporogenes.
IPA metabolism diagram
^ "3-Indolepropionic acid". Human Metabolome Database. University of Alberta. Retrieved 12 June 2018.Indole-3-propionate (IPA), a deamination product of tryptophan formed by symbiotic bacteria in the gastrointestinal tract of mammals and birds. 3-Indolepropionic acid has been shown to prevent oxidative stress and death of primary neurons and neuroblastoma cells exposed to the amyloid beta-protein in the form of amyloid fibrils, one of the most prominent neuropathologic features of Alzheimer's disease. 3-Indolepropionic acid also shows a strong level of neuroprotection in two other paradigms of oxidative stress. (
PMID 10419516) ... More recently it has been found that higher indole-3-propionic acid levels in serum/plasma are associated with reduced likelihood of type 2 diabetes and with higher levels of consumption of fiber-rich foods (
PMID 28397877)
Origin: • Endogenous • Microbial
^ Chyan YJ, Poeggeler B, Omar RA, Chain DG, Frangione B, Ghiso J, Pappolla MA (July 1999). "Potent neuroprotective properties against the Alzheimer beta-amyloid by an endogenous melatonin-related indole structure, indole-3-propionic acid". J. Biol. Chem. 274 (31): 21937–21942. doi:10.1074/jbc.274.31.21937. PMID 10419516.[Indole-3-propionic acid (IPA)] has previously been identified in the plasma and cerebrospinal fluid of humans, but its functions are not known. ... In kinetic competition experiments using free radical-trapping agents, the capacity of IPA to scavenge hydroxyl radicals exceeded that of melatonin, an indoleamine considered to be the most potent naturally occurring scavenger of free radicals. In contrast with other antioxidants, IPA was not converted to reactive intermediates with pro-oxidant activity.
^ Mendes-Soares H, Suzuki H, Hickey RJ, Forney LJ (April 2014). "Comparative functional genomics of Lactobacillus spp. reveals possible mechanisms for specialization of vaginal lactobacilli to their environment". Journal of Bacteriology. 196 (7): 1458–70. doi:10.1128/JB.01439-13. PMC 3993339. PMID 24488312.
^ Sun Z, Harris HM, McCann A, Guo C, Argimón S, Zhang W, Yang X, Jeffery IB, Cooney JC, Kagawa TF, Liu W, Song Y, Salvetti E, Wrobel A, Rasinkangas P, Parkhill J, Rea MC, O'Sullivan O, Ritari J, Douillard FP, Paul Ross R, Yang R, Briner AE, Felis GE, de Vos WM, Barrangou R, Klaenhammer TR, Caufield PW, Cui Y, Zhang H, O'Toole PW (September 2015). "Expanding the biotechnology potential of lactobacilli through comparative genomics of 213 strains and associated genera". Nature Communications. 6 (1): 8322. Bibcode:2015NatCo...6E8322S. doi:10.1038/ncomms9322. PMC 4667430. PMID 26415554.
^ France MT, Mendes-Soares H, Forney LJ (December 2016). "Genomic Comparisons of Lactobacillus crispatus and Lactobacillus iners Reveal Potential Ecological Drivers of Community Composition in the Vagina". Applied and Environmental Microbiology. 82 (24): 7063–7073. doi:10.1128/AEM.02385-16. PMC 5118917. PMID 27694231.
^ Basharat Z, Yasmin A (December 2015). "Survey of compound microsatellites in multiple Lactobacillus genomes". Canadian Journal of Microbiology. 61 (12): 898–902. doi:10.1139/cjm-2015-0136. hdl:1807/69860. PMID 26445296.
^ "Archived copy". Archived from the original on 2007-02-02. Retrieved 2007-02-02.CS1 maint: Archived copy as title (link)
^ (IJSEM, Paper in Press).
^ ab Wang ZK, Yang YS, Stefka AT, Sun G, Peng LH (April 2014). "Review article: fungal microbiota and digestive diseases". Alimentary Pharmacology & Therapeutics. 39 (8): 751–66. doi:10.1111/apt.12665. PMID 24612332.In addition, GI fungal infection is reported even among those patients with normal immune status. Digestive system-related fungal infections may be induced by both commensal opportunistic fungi and exogenous pathogenic fungi. ...
In vitro, bacterial hydrogen peroxide or organic acids can inhibit C. albicans growth and virulence61
In vivo, Lactobacillus sp. can inhibit the GI colonisation and infection of C. albicans62
In vivo, C. albicans can suppress Lactobacillus sp. regeneration in the GI tract after antibiotic therapy63, 64
^ ab Erdogan A, Rao SS (April 2015). "Small intestinal fungal overgrowth". Current Gastroenterology Reports. 17 (4): 16. doi:10.1007/s11894-015-0436-2. PMID 25786900.Small intestinal fungal overgrowth (SIFO) is characterized by the presence of excessive number of fungal organisms in the small intestine associated with gastrointestinal (GI) symptoms. Candidiasis is known to cause GI symptoms particularly in immunocompromised patients or those receiving steroids or antibiotics. However, only recently, there is emerging literature that an overgrowth of fungus in the small intestine of non-immunocompromised subjects may cause unexplained GI symptoms. ... Fungal-bacterial interaction may act in different ways and may either be synergistic or antagonistic or symbiotic [29]. Some bacteria such as Lactobacillus species can interact and inhibit both the virulence and growth of Candida species in the gut by producing hydrogen peroxide [30]. Any damage to the mucosal barrier or disruption of GI microbiota with chemotherapy or antibiotic use, inflammatory processes, activation of immune molecules and disruption of epithelial repair may all cause fungal overgrowth [27].
^ abcd Vilela SF, Barbosa JO, Rossoni RD, Santos JD, Prata MC, Anbinder AL, Jorge AO, Junqueira JC (February 2015). "Lactobacillus acidophilus ATCC 4356 inhibits biofilm formation by C. albicans and attenuates the experimental candidiasis in Galleria mellonella". Virulence. 6 (1): 29–39. doi:10.4161/21505594.2014.981486. PMC 4603435. PMID 25654408.
^ Axelsson, L. T.; Chung, T. C.; Dobrogosz, W. J.; Lindgren, S. E. (April 1988). "Production of a Broad Spectrum Antimicrobial Substance by Lactobacillus reuteri". Microbial Ecology in Health and Disease. 2 (2): 131–136. doi:10.3109/08910608909140210.
^ Brink, B. ten; Minekus, M.; van der Vossen, J.M.B.M.; Leer, R.J.; Huis in't Veld, J.H.J. (August 1994). "Antimicrobial activity of lactobacilli: preliminary characterization and optimization of production of acidocin B, a novel bacteriocin produced by Lactobacillus acidophilus M46". Journal of Applied Microbiology. 77 (2): 140–148. doi:10.1111/j.1365-2672.1994.tb03057.x.
^ Ford AC, Quigley EM, Lacy BE, Lembo AJ, Saito YA, Schiller LR, Soffer EE, Spiegel BM, Moayyedi P (October 2014). "Efficacy of prebiotics, probiotics, and synbiotics in irritable bowel syndrome and chronic idiopathic constipation: systematic review and meta-analysis". The American Journal of Gastroenterology. 109 (10): 1547–61, quiz 1546, 1562. doi:10.1038/ajg.2014.202. PMID 25070051.
^ abc Ruggiero P (November 2014). "Use of probiotics in the fight against Helicobacter pylori". World Journal of Gastrointestinal Pathophysiology. 5 (4): 384–91. doi:10.4291/wjgp.v5.i4.384. PMC 4231502. PMID 25400981.
^ ab Cribby S, Taylor M, Reid G (March 9, 2009). "Vaginal microbiota and the use of probiotics". Interdisciplinary Perspectives on Infectious Diseases. 2008: 256490. doi:10.1155/2008/256490. PMC 2662373. PMID 19343185.
^ Ashraf, Shah, Rabia, Nagendra P (2014). "Immune system stimulation by probiotic microorganisms". Critical Reviews in Food Science and Nutrition. 54 (7): 938–56. doi:10.1080/10408398.2011.619671. PMID 24499072.
^ Twetman S, Stecksén-Blicks C (January 2008). "Probiotics and oral health effects in children". International Journal of Paediatric Dentistry. 18 (1): 3–10. doi:10.1111/j.1365-263X.2007.00885.x. PMID 18086020.
^ Meurman JH, Stamatova I (September 2007). "Probiotics: contributions to oral health". Oral Diseases. 13 (5): 443–51. doi:10.1111/j.1601-0825.2007.01386.x. PMID 17714346.
^ abcd Grenier, Daniel; et al. (October 2009). "Probiotics for Oral Health: Myth or Reality?" (PDF). Professional Issues. 75 (8): 585–590 – via Google.
^ Inglin RC, Stevens MJ, Meile L, Lacroix C, Meile L (July 2015). "High-throughput screening assays for antibacterial and antifungal activities of Lactobacillus species". Journal of Microbiological Methods. 114 (July 2015): 26–9. doi:10.1016/j.mimet.2015.04.011. PMID 25937247.
^ Inglin, Raffael (2017). "PhD Thesis - Combined Phenotypic-Genotypic Analyses of the Genus Lactobacillus and Selection of Cultures for Biopreservation of Fermented Food". ETH Zurich. doi:10.3929/ethz-b-000214904.
External links[edit]
 Data related to Lactobacillus at Wikispecies
Data related to Lactobacillus at Wikispecies- List of species of the genus Lactobacillus
- Lactobacillus at Milk the Funk Wiki
Lactobacillus at BacDive - the Bacterial Diversity Metadatabase
Categories:
- Lactobacillaceae
- Food science
- Gut flora bacteria
- Garde manger
- Gram-positive bacteria
(window.RLQ=window.RLQ||).push(function(){mw.config.set({"wgPageParseReport":{"limitreport":{"cputime":"1.312","walltime":"1.579","ppvisitednodes":{"value":6705,"limit":1000000},"ppgeneratednodes":{"value":0,"limit":1500000},"postexpandincludesize":{"value":325253,"limit":2097152},"templateargumentsize":{"value":44639,"limit":2097152},"expansiondepth":{"value":14,"limit":40},"expensivefunctioncount":{"value":18,"limit":500},"unstrip-depth":{"value":1,"limit":20},"unstrip-size":{"value":129360,"limit":5000000},"entityaccesscount":{"value":14,"limit":400},"timingprofile":["100.00% 1159.640 1 -total"," 44.44% 515.383 1 Template:Reflist"," 27.93% 323.881 29 Template:Cite_journal"," 16.25% 188.391 1 Template:Automatic_taxobox"," 11.69% 135.608 15 Template:Navbox"," 9.70% 112.477 3 Template:Cite_web"," 8.66% 100.370 2 Template:More_citations_needed"," 7.91% 91.671 1 Template:Taxonbar"," 6.83% 79.211 2 Template:Ambox"," 6.03% 69.965 1 Template:Tryptophan_metabolism_by_human_microbiota"]},"scribunto":{"limitreport-timeusage":{"value":"0.705","limit":"10.000"},"limitreport-memusage":{"value":7946351,"limit":52428800},"limitreport-logs":"table#1 {n ["size"] = "tiny",n}n"},"cachereport":{"origin":"mw1271","timestamp":"20190313224242","ttl":2592000,"transientcontent":false}}});});{"@context":"https://schema.org","@type":"Article","name":"Lactobacillus","url":"https://en.wikipedia.org/wiki/Lactobacillus","sameAs":"http://www.wikidata.org/entity/Q1061596","mainEntity":"http://www.wikidata.org/entity/Q1061596","author":{"@type":"Organization","name":"Contributors to Wikimedia projects"},"publisher":{"@type":"Organization","name":"Wikimedia Foundation, Inc.","logo":{"@type":"ImageObject","url":"https://www.wikimedia.org/static/images/wmf-hor-googpub.png"}},"datePublished":"2003-02-28T04:54:45Z","dateModified":"2019-03-12T05:38:21Z","image":"https://upload.wikimedia.org/wikipedia/commons/7/7d/Lactobacillus_sp_01.png","headline":"genus of bacteria"}(window.RLQ=window.RLQ||).push(function(){mw.config.set({"wgBackendResponseTime":288,"wgHostname":"mw1238"});});